Differential Scanning Fluorometry (nanoDSF)
nanoDSF is a biophysical technique used to assess the conformational stability of protein, which essentially means it checks how likely a protein is to remain folded and functional under stressful conditions. It works by monitoring a protein’s intrinsic fluorescence as it responds to thermal or chemical stress. As the protein unfolds under this stress, its fluorescence properties change. The point at which unfolding occurs (“melting temperature”) is indicative of the overall conformational stability of the protein.
nanoDSF can detect even minor changes in a protein's fluorescence, providing a very detailed picture of its conformational stability. It can measure both thermal stability (how a protein behaves with increasing temperature) and chemical stability (how it behaves in the presence of denaturing chemicals).
Moreover, nanoDSF can also record even very small changes of protein conformational stability coming from binding of a ligand. This can be used for efficient, high-throughput screening for ligand binding (Thermal Shift Assay, TSA). Importantly, since nanoDSF works with intrinsic protein fluorescence, neither fluorescent labeling, nor the presence of an external fluorescent dye are necessary.
Deduced Parameters
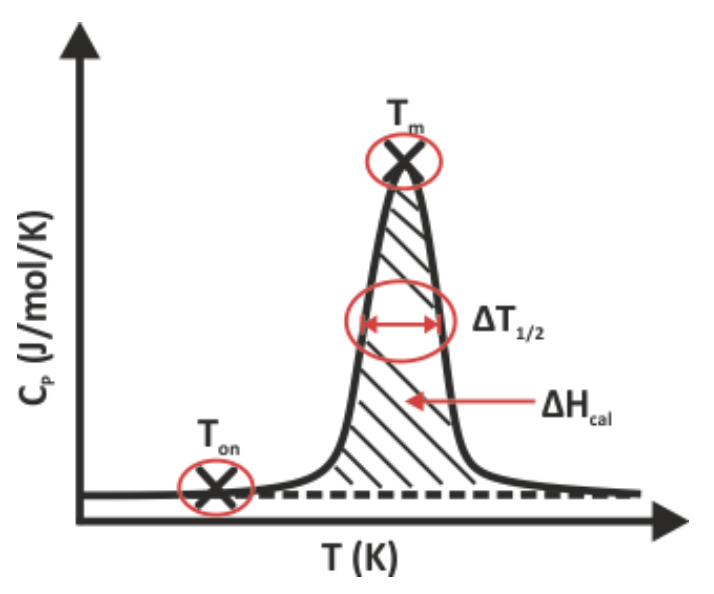
Tm - Melting temperature; the temperature at which half of a protein sample is unfolded
Ton - Onset temperature; the temperature at which protein unfolding starts
EA - Activation energy of unfolding
Ton-turbidity - Onset temperature of aggregation; the temperature at which protein aggregates start to appear
Tm-diff - Difference of melting temperature of a protein in presence and absence of a ligand (e.g. small molecule)
EC50 -Dose-response format of Tm-diff measurement
Wir benötigen Ihre Zustimmung zum Laden der Übersetzungen
Wir nutzen einen Drittanbieter-Service, um den Inhalt der Website zu übersetzen, der möglicherweise Daten über Ihre Aktivitäten sammelt. Bitte überprüfen Sie die Details in der Datenschutzerklärung und akzeptieren Sie den Dienst, um die Übersetzungen zu sehen.