Wernig-Zorc, S. et al.; Sci. Adv. 10, (2024); pmid: 38959309
nucMACC: An MNase-seq pipeline to identify structurally altered nucleosomes in the genome.
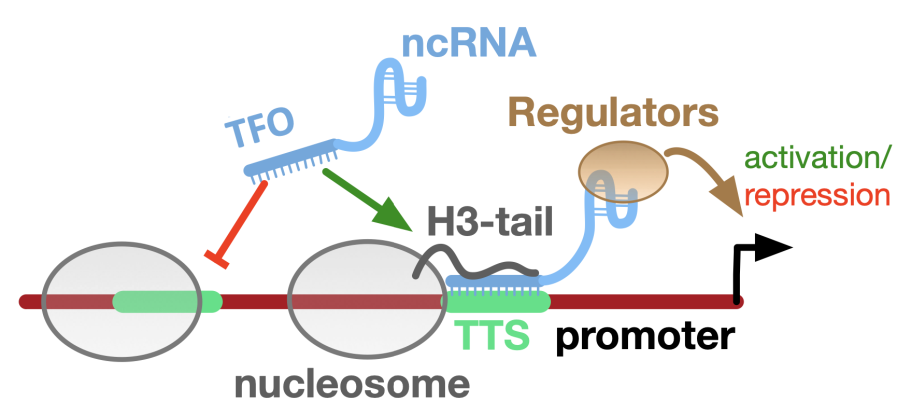
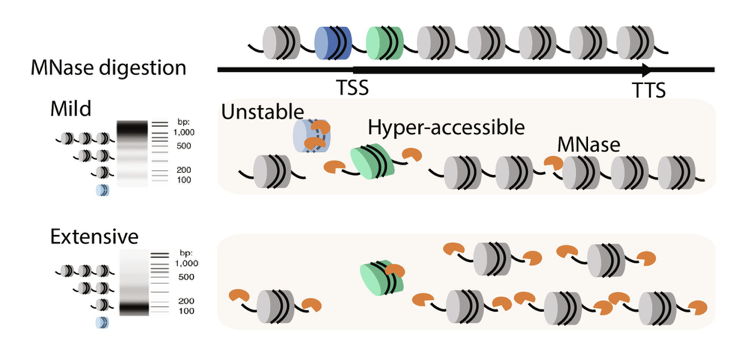
We present the nucleosome-based MNase accessibility (nucMACC) pipeline unveiling the regulatory chromatin landscape by measuring nucleosome accessibility and stability. The nucMACC pipeline represents a systematic and genome-wide approach for detecting unstable (“fragile”) nucleosomes.
We have characterized the regulatory nucleosome landscape in Drosophila melanogaster, Saccharomyces cerevisiae, and mammals.
Two functionally distinct sets of promoters were identified, one associated with an unstable nucleosome and the other being nucleosome depleted. We show that unstable nucleosomes present intermediate states of nucleosome remodeling, preparing inducible genes for transcriptional activation in response to stimuli or stress. The presence of unstable nucleosomes correlates with RNA polymerase II proximal pausing.
The nucMACC pipeline offers unparalleled precision and depth in nucleosome research and is a valuable tool for future nucleosome studies.
2024 - A new bioinformatic pipeline to reveal cellular chromatin architecture
2023 - Analysis of the nuclear localisation signals of Chd4-NuRD
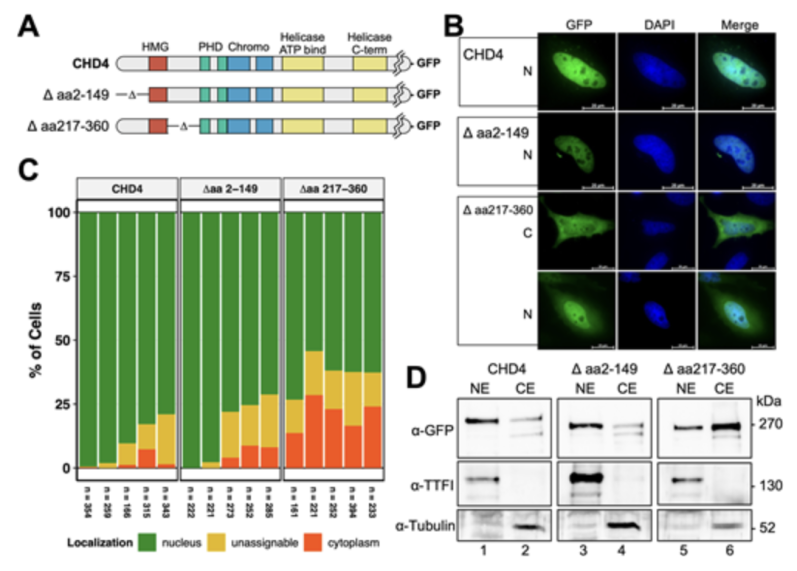
Chromatin remodeling enzymes form large multiprotein complexes that play central roles inregulating access to the genome. Here, we characterize the nuclear import of the humanCHD4 protein. We show that CHD4 enters the nucleus by means of several importin alphaproteins (1, 5, 6, and 7), and rather independently of importin beta 1. Importin alpha 1 directlyinteracts with a monopartite “KRKR”-motif in the N-terminus of CHD4 (aa 304-307).However, alanine mutagenesis of this motif only leads to approx. 50% reduction in nuclearlocalization of CHD4, implying additional import mechanisms. Interestingly, we can showthat CHD4 is already associated with NuRD core subunits such as MTA2, HDAC1, orRbAp46 in the cytoplasm, suggesting an assembly of the NuRD core complex before nuclearimport. We propose that, in addition to the importin-alpha-dependent nuclear localizationsignal, CHD4 is dragged into the nucleus by a "piggyback mechanism" using the importsignals of the associated NuRD subunits.
2023 - Analysing Adenovirus chromatin changes during early infection
Unpacking the smart way; How Incoming adenoviruses change their chromatin structure for efficient gene expression
Adenoviruses, known for their low pathogenicity and technological approachability, have become instrumental in many therapeutic applications, including as vaccination vector platform during the recent SARS-CoV-2 pandemic. Central to their efficacy is the remarkably efficient delivery and expression of their genetic material.
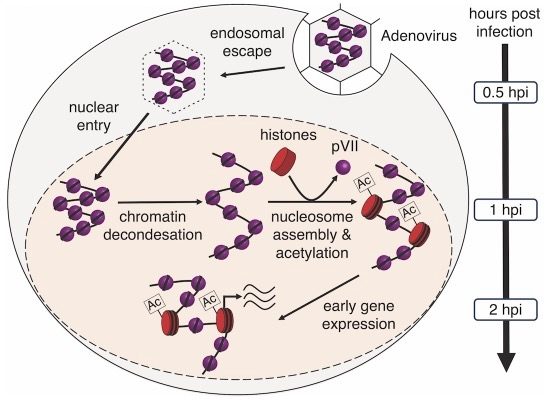
Two scientific teams led by Gernot Längst from the University of Regensburg and Harald Wodrich from the University of Bordeaux have obtained insight into this process in unprecedented detail. Employing advanced microscopy techniques and genome structure analyses, they traced the early stages of adenoviral infection in human cells. This approach allowed a detailed description of the viral genome intricately wrapped with the viral pVII protein within the incoming virion. Furthermore, their investigation revealed the transformation of the viral chromatin structure, converting the densely packed genome into an exceptionally efficient template for gene expression as it traverses from the cell surface to the cell nucleus.
The scientist could show that in the virus, the viral DNA is tightly wrapped around the viral pVII protein. This unique conformation facilitates seamless accommodation of the genome within the confined viral capsid. Once viruses entered the cell, the packaged viral DNA completes its journey to the nucleus in approximately 30 minutes. Following capsid release and nuclear import parts of the viral genome alter the transport configuration and open up. During this process, a few pVII proteins are specifically removed and replaced by cellular histone proteins at the regulatory sites of early activated viral genes. This strategic exchange with histone proteins unlocks the potential for the viral genetic information to hijack the cellular transcription machinery, enabling efficient gene expression – a process that starts around thirty minutes after the genome arrives in the nucleus, culminating in cell infection.
By comprehending the intricate structural dynamics of the adenoviral genome during the initial stages of infection, the researchers aim to leverage this knowledge for the enhancement of adenoviral vector design. This holds promise for refining applications such as vaccination and gene therapy, thereby elevating both safety and efficiency.
The study was recently published in the Journal of the European Molecular Biology Organization (EMBO Journal).
Wir benötigen Ihre Zustimmung zum Laden der Übersetzungen
Wir nutzen einen Drittanbieter-Service, um den Inhalt der Website zu übersetzen, der möglicherweise Daten über Ihre Aktivitäten sammelt. Bitte überprüfen Sie die Details in der Datenschutzerklärung und akzeptieren Sie den Dienst, um die Übersetzungen zu sehen.