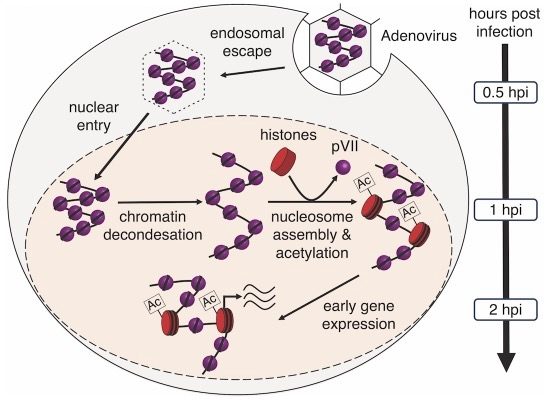
Adenoviruses
Adenoviruses are non-enveloped viruses characterized by a icosahedral capsid structure and a complex nucleoprotein organization.
Inside the capsid, the viral core contains the adenovirus genome bound to protein pVII (most abundant DA binding protein), pV and Mu(pX). These proteins are rich in basic amino acids, enabling them to bind tightly to the negatively charged viral DNA. This binding condenses the DNA into a compact nucleoprotein structure, allowing the DNA to fit into the limited space of the capsid.The virus has to dramatically re-organize its DNA packaging to allow transcription and replication in the host cell.
We are interested in how the DNA packaging of the virus is organized and changes during the infection cycle.
We apply high resolution microscopy and genomics technologies to uncover changes in viral DNA packaging during its life cycle and track its associated gene expression activity.
Corona Virus
Our research focuses on the multifaceted roles of the SARS-CoV-2 nucleocapsid (N) protein, a central player in the viral replication cycle and host-virus interactions. The N protein is essential for viral RNA packaging, genome stability, and replication, as well as the regulation of host immune responses. Its ability to bind and modulate viral RNA is key to the proper assembly and functioning of the viral genome. The N protein is composed of structured domains, including the RNA-binding domain (RBD) and C-terminal domain (CTD), along with several intrinsically disordered regions (IDRs) that contribute to its functional diversity.
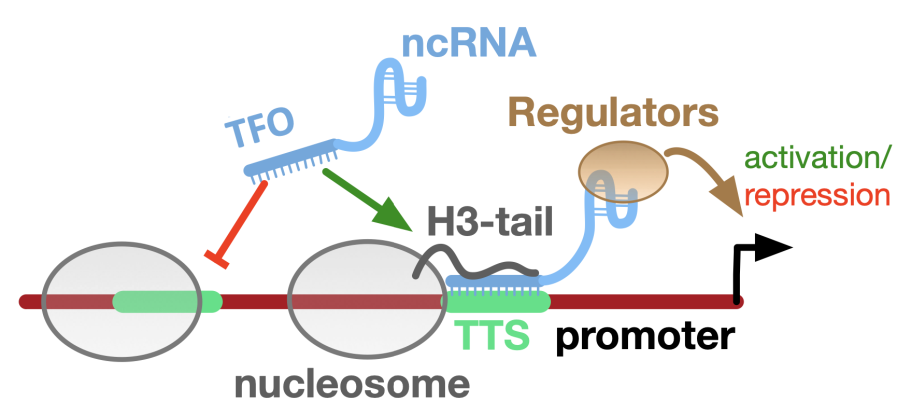
Triple-Helices
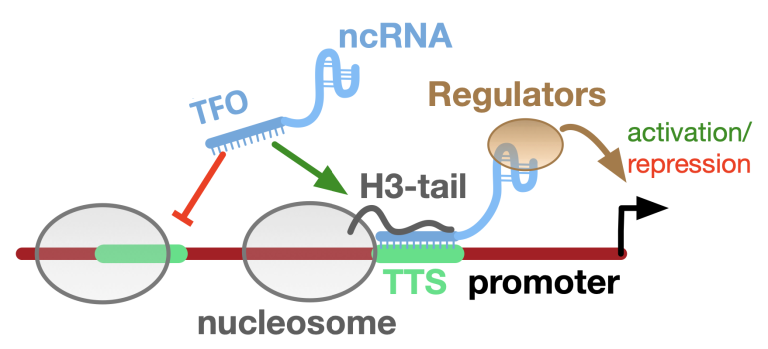
RNA-DNA triple helices play a role in Gene Regulation and Chromatin Structure. Triplexes can regulate gene expression by forming at promoters or enhancers, influencing transcription factor binding and chromatin structure, altering chromatin accessibility and impacting gene expression during development and differentiation.
The triplex structures are sequence-specific and less stable than Watson-Crick pairings, but they are stabilized by the histone H3 tail of neighboring nucleosomes. We are studying the Triplex-Chromatin interaction and its application in the regulation of gene expression.
Plasmodium falciparum Chromatin
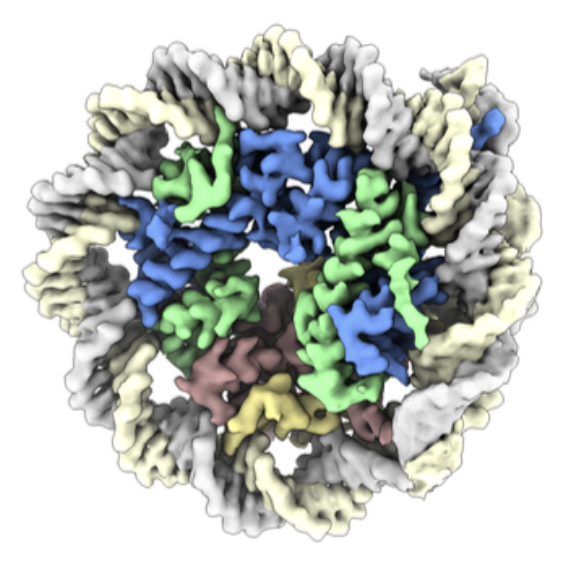
Malaria, caused by Plasmodium falciparum (Pf) is one of the biggest threats to human health. Pf is an highly unusual eukaryote, with an extraordinarily high AT-content (>80%) of its genome and the most distinct histone and CRE proteins among all eukaryotes. We are interested in how functional chromatin is established in the non-flexible AT-rich DNA and how epigenetic mechanisms contribute to the regulation of the complex life cycle of this parasite.
Chromatin Remodeler
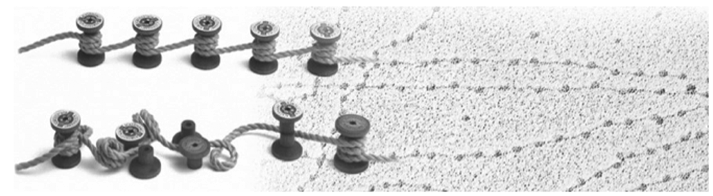
Chromatin remodeling enzymes (CRE) are ATP dependent, multiprotein complexes that move or disrupt histone octamers on DNA, or exchange specific histone proteins within the octameric particle. We aim to study the mechanisms and regulation of the CREs, adressing the molecular mechanism of nucleosome positioning and their regulation by non-coding RNAs.
Higher Order Structure of Chromatin

The textbook illustration of higher order structures of chromatin, depicting a hierarchical and regular folding of the nucleosomal array to the compacted chromosome, does most probably not exist in cells. We suggest that cellular chromatin exist as a dynamic, but irregular structure, following the polymer melt model. Our aim is to unravel the nuclear architecture and organisation of chromatin in cells.
Wir benötigen Ihre Zustimmung zum Laden der Übersetzungen
Wir nutzen einen Drittanbieter-Service, um den Inhalt der Website zu übersetzen, der möglicherweise Daten über Ihre Aktivitäten sammelt. Bitte überprüfen Sie die Details in der Datenschutzerklärung und akzeptieren Sie den Dienst, um die Übersetzungen zu sehen.