Chromatin Research in our Lab
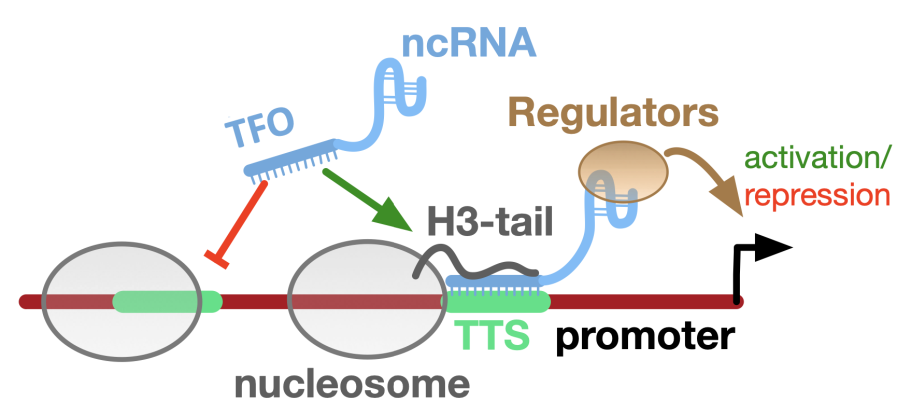
Chromatin remodeling enzymes (CRE) are ATP dependent, multiprotein complexes that move or disrupt histone octamers on DNA, or exchange specific histone proteins within the octameric particle. We aim to study the mechanisms and regulation of the CREs, adressing the molecular mechanism of nucleosome positioning and their regulation by non-coding RNAs.
Chromatin Dynamics
Higher Order Structures of Chromatin
The textbook illustration of higher order structures of chromatin, depicting a hierarchical and regular folding of the nucleosomal array to the compacted chromosome, does most probably not exist in cells. We suggest that cellular chromatin exist as a dynamic, but irregular structure, following the polymer melt model. Our aim is to unravel the nuclear architecture and organisation of chromatin in cells.
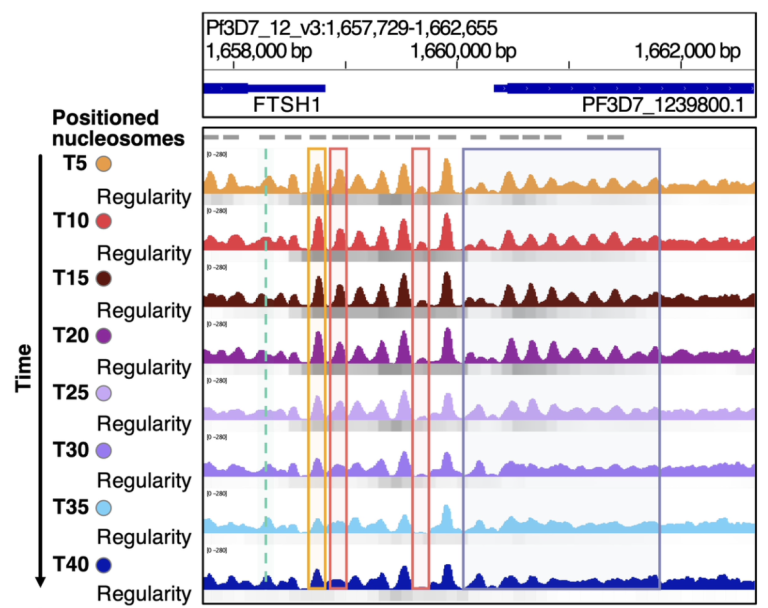
Chromatin Dynamics in Plasmodium falciparum
Malaria, caused by Plasmodium falciparum (Pf) is one of the biggest threats to human health. Pf is an highly unusual eukaryote, with an extraordinarily high AT-content (>80%) of its genome and the most distinct histone and CRE proteins among all eukaryotes. We are interested in how functional chromatin is established in the non-flexible AT-rich DNA and how epigenetic mechanisms contribute to the regulation of the complex life cycle of this parasite.
Mechanisms of nucleosome positioning
Nucleosome positioning on DNA determines the accessibility of regulatory elements to their cognate transcription factors and is regulated by CREs and the intrinsic positioning of histone octamers that is influenced by the sequence dependent DNA structure. We study how CREs read the signals of DNA sequence/structure to position nucleosomes and address the role of histone tails in selecting the intrinsic positions of histone octamers on genomic DNA.
Wir benötigen Ihre Zustimmung zum Laden der Übersetzungen
Wir nutzen einen Drittanbieter-Service, um den Inhalt der Website zu übersetzen, der möglicherweise Daten über Ihre Aktivitäten sammelt. Bitte überprüfen Sie die Details in der Datenschutzerklärung und akzeptieren Sie den Dienst, um die Übersetzungen zu sehen.